Non-referenced genome assembly from epigenomic short-read data.
Antony Kaspi, Mark Ziemann, Samuel T Keating, Ishant Khurana, Timothy Connor, Briana Spolding, Adrian Cooper, Ross Lazarus, Ken Walder, Paul Zimmet, Assam El-Osta
文献索引:Epigenetics 9(10) , 1329-38, (2014)
全文:HTML全文
摘要
Current computational methods used to analyze changes in DNA methylation and chromatin modification rely on sequenced genomes. Here we describe a pipeline for the detection of these changes from short-read sequence data that does not require a reference genome. Open source software packages were used for sequence assembly, alignment, and measurement of differential enrichment. The method was evaluated by comparing results with reference-based results showing a strong correlation between chromatin modification and gene expression. We then used our de novo sequence assembly to build the DNA methylation profile for the non-referenced Psammomys obesus genome. The pipeline described uses open source software for fast annotation and visualization of unreferenced genomic regions from short-read data.
相关化合物
| 结构式 | 名称/CAS号 | 分子式 | 全部文献 |
|---|---|---|---|
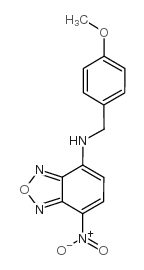 |
7-(对甲氧基苄氨基)-4-硝基苯-2-恶唑-1,3-二唑
CAS:33984-50-8 |
C14H12N4O4 |
|
Coordinated epigenetic remodelling of transcriptional networ...
2015-01-01 [Clin. Epigenetics. 7 , 52, (2015)] |
|
Impact of the F508del mutation on ovine CFTR, a Cl- channel ...
2015-06-01 [J. Physiol. 593 , 2427-46, (2015)] |
|
Nutritional evaluation of Australian microalgae as potential...
2015-01-01 [PLoS ONE 10(2) , e0118985, (2015)] |
|
Electrochemical detection of anti-tau antibodies binding to ...
2016-03-01 [Anal. Biochem. 496 , 55-62, (2016)] |
|
Targeting of P-Element Reporters to Heterochromatic Domains ...
2016-02-01 [Genetics 202 , 565-82, (2016)] |