The structure of recA protein-DNA filaments. 2 recA protein monomers unwind 17 base pairs of DNA by 11.5 degrees/base pair in the presence of adenosine 5'-O-(3-thiotriphosphate).
S Chrysogelos, J C Register, J Griffith
文献索引:J. Biol. Chem. 258 , 12624, (1983)
全文:HTML全文
摘要
recA protein binds to duplex DNA in the presence of Mg2+ and adenosine 5'-O-(3-thiotriphosphate) forming a stiff nucleoprotein filament with a distinct axial repeat which contains 17 +/- 1 base pairs and spans 8-9 nm along the fiber (Di Capua, E., Engel, A., Stasiak, A., and Koller, Th. (1982) J. Mol. Biol. 157, 87-103; Dunn, K., Chrysogelos, S., and Griffith, J. (1982) Cell 28, 757-765). Measurement of the protein:DNA ratio in these filaments utilizing double label analysis and isopycnic density banding shows that there are 2 recA monomers for every 17 base pairs. The DNA is also partially unwound in this filament. Utilizing the recA-induced relaxation of naturally supertwisted SV40 DNA, we show that the DNA is unwound by 11.5 +/- 1.5 degrees/base pair which corresponds to 180-200 degrees for each repeat unit along the filament length.
相关化合物
| 结构式 | 名称/CAS号 | 分子式 | 全部文献 |
|---|---|---|---|
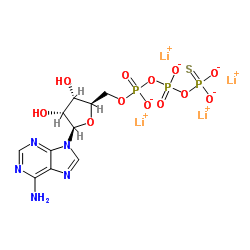 |
Adenosine 5'-(γ-thio)-triphosphate (lithium salt)
CAS:93839-89-5 |
C10H12Li4N5O12P3S |
|
Specific inhibition of p97/VCP ATPase and kinetic analysis d...
2014-07-29 [J. Mol. Biol. 426(15) , 2886-99, (2014)] |
|
The enzymatic synthesis of thiophosphate analogs of nucleoti...
1972-07-13 [Biochim. Biophys. Acta 276 , 155, (1972)] |
|
ATP binds to proteasomal ATPases in pairs with distinct func...
2011-02-18 [Cell 144 , 526-538, (2011)] |
|
ATP-Dependent Steps in the Binding of Ubiquitin Conjugates t...
2010-11-24 [Mol. Cell. 40 , 671-781, (2010)] |
|
Extracellular ATP differentially modulates Toll-like recepto...
2011-03-01 [J. Neurochem. 116 , 1138-1147, (2011)] |