Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants.
M M Santoro, D W Bolen
文献索引:Biochemistry 27(21) , 8063-8, (1988)
全文:HTML全文
摘要
Characteristics and properties of the unfolding free energy change, delta G degrees N-U, as determined by the linear extrapolation method are assessed for the unfolding of phenylmethanesulfonyl chymotrypsin (PMS-Ct). Difference spectral measurements at 293 nm were used to define PMS-Ct unfolding brought about with guanidinium chloride, urea, and 1,3-dimethylurea. All three denaturants were shown to give identical extinction coefficient differences (delta epsilon N-U) between native and unfolded forms of the protein in the limit of zero concentration of denaturant. The independence of delta epsilon N-U on denaturant supports the linear extension of pre- and postdenaturational base lines into the transition zone, allowing evaluation of unfolding equilibrium constants based on the two-state assumption. An expression, based on the linear extrapolation method, was used to provide estimates of delta G degrees N-U for the three denaturants using nonlinear least-squares fitting of the primary data, delta epsilon versus [denaturant]. The three delta G degrees N-U values were identical, within error, suggesting that the free energy change is a property of the protein system and independent of denaturant. It is suggested that the error in delta G degrees N-U determined from use of the linear extrapolation method is significantly larger than commonly reported in the literature.
相关化合物
| 结构式 | 名称/CAS号 | 分子式 | 全部文献 |
|---|---|---|---|
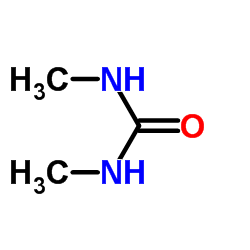 |
1,3-二甲基脲
CAS:96-31-1 |
C3H8N2O |
|
Differential protein abundance and function of UT-B urea tra...
2010-03-01 [Am. J. Physiol. Gastrointest. Liver Physiol. 298(3) , G345-51, (2010)] |
|
In vitro stimulation by paraquat of reactive oxygen-mediated...
1981-09-15 [Toxicol. Appl. Pharmacol. 60(2) , 279-86, (1981)] |
|
Novel bUT-B2 urea transporter isoform is constitutively acti...
2009-08-01 [Am. J. Physiol. Regul. Integr. Comp. Physiol. 297(2) , R323-9, (2009)] |
|
Acute regulation of mUT-A3 urea transporter expressed in a M...
2007-04-01 [Am. J. Physiol. Renal Physiol. 292(4) , F1157-63, (2007)] |
|
Multiple hydrogen-bonding interactions between macrocyclic t...
2011-04-28 [Phys. Chem. Chem. Phys. 13(16) , 7384-95, (2011)] |