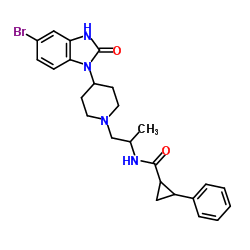
1246303-14-9
| Name | (1R,2R)-N-{(2S)-1-[4-(5-Bromo-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)-1-piperidinyl]-2-propanyl}-2-phenylcyclopropanecarboxamide |
|---|---|
| Synonyms | (1R,2R)-N-{(2S)-1-[4-(5-Bromo-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)-1-piperidinyl]-2-propanyl}-2-phenylcyclopropanecarboxamide |
| Description | VU0359595 (CID-53361951; ML-270) is a potent and selective pharmacological phospholipase D1 (PLD1) inhibitor with an IC50 of 3.7 nM. VU0359595 is >1700-fold selective for PLD1 over PLD2 (IC50 of 6.4 μM). VU0359595 can be used for the research of cancer, diabetes, neurodegenerative and inflammatory diseases[1][2][3][4]. |
|---|---|
| Related Catalog | |
| Target |
PLD1:3.7 nM (IC50) PLD2:6.4 μM (IC50) |
| In Vitro | VU0359595 (5, 50, 500, 5000 nM) inhibits basal and FCS/IGF-1 stimulated proliferation of astroglial cells[2] . VU0359595 (5, 50, 500 nM; 30 min) does not affect basal PLD activity in astrocytes but reduces mitogen-stimulated PLD activity in a concentration-dependent manner[2]. VU0359595 (0.15 μM; 1 h before high glucose treatment and 4 h during high glucose treatment) partially reduces the increase [3H]-phosphatidylethanol (PEth) generation induced by high glucose (33 mM) in retinal pigment epithelium (RPE) cells[3]. VU0359595 (5 μM; 1 h prior to LPS treatment) modulates the autophagic process of LPS-induced (10 μg/ml; 24 h) RPE cells[4]. |
| References |
| Density | 1.4±0.1 g/cm3 |
|---|---|
| Molecular Formula | C25H29BrN4O2 |
| Molecular Weight | 497.427 |
| Exact Mass | 496.147369 |
| LogP | 4.71 |
| Index of Refraction | 1.631 |
| RIDADR | NONH for all modes of transport |
|---|