| Structure | Name/CAS No. | Articles |
|---|---|---|
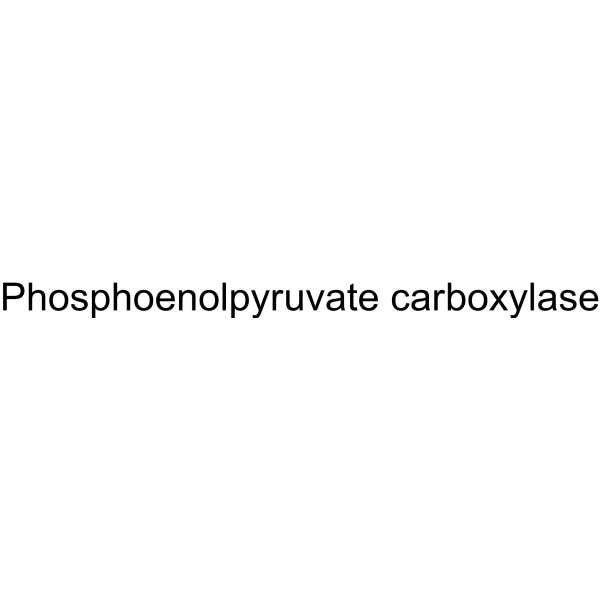 |
Phosphoenolpyruvate carboxylase
CAS:9067-77-0 |
|
 |
Malate dehydrogenase oxaloacetate-decarboxylating, NADP+
CAS:9028-47-1 |