Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Jia Sheng, Wen Zhang, Abdalla E A Hassan, Jianhua Gan, Alexei S Soares, Song Geng, Yi Ren, Zhen Huang
Index: Nucleic Acids Res. 40(16) , 8111-8, (2012)
Full Text: HTML
Abstract
Natural RNAs, especially tRNAs, are extensively modified to tailor structure and function diversities. Uracil is the most modified nucleobase among all natural nucleobases. Interestingly, >76% of uracil modifications are located on its 5-position. We have investigated the natural 5-methoxy (5-O-CH(3)) modification of uracil in the context of A-form oligonucleotide duplex. Our X-ray crystal structure indicates first a H-bond formation between the uracil 5-O-CH(3) and its 5'-phosphate. This novel H-bond is not observed when the oxygen of 5-O-CH(3) is replaced with a larger atom (selenium or sulfur). The 5-O-CH(3) modification does not cause significant structure and stability alterations. Moreover, our computational study is consistent with the experimental observation. The investigation on the uracil 5-position demonstrates the importance of this RNA modification at the atomic level. Our finding suggests a general interaction between the nucleobase and backbone and reveals a plausible function of the tRNA 5-O-CH(3) modification, which might potentially rigidify the local conformation and facilitates translation.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
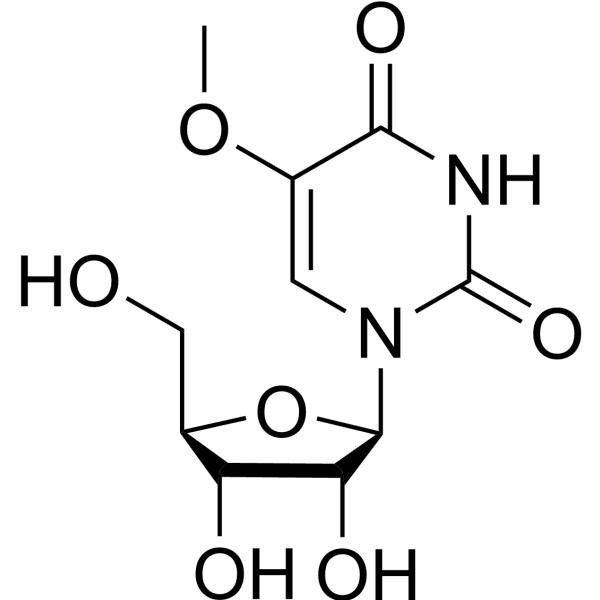 |
5-methoxyuridine
CAS:35542-01-9 |
C10H14N2O7 |
|
Codon recognition by tRNA molecules with a modified or unmod...
1995-01-01 [Nucleic Acids Symp. Ser. 34 , 203-204, (1995)] |
|
5-methoxyuridine: a new minor constituent located in the fir...
1976-10-01 [Nucleic Acids Res. 3 , 2851-2860, (1976)] |
|
Bacillus subtilis tRNA(Pro) with the anticodon mo5UGG can re...
2005-05-01 [Biochim. Biophys. Acta 1728 , 143-149, (2005)] |
|
5-Methoxyuridine, a new modified constituent in tRNAs of Bac...
1976-11-01 [FEBS Lett. 70 , 37-42, (1976)] |
|
Nucleotide sequence of valine tRNA mo5UAC from bacillus subt...
1982-01-22 [Nucleic Acids Res. 10(2) , 715-8, (1982)] |