DNA damage induced in vivo by 7-methoxy-2-nitronaphtho[2,1-b]-furan (R7000) in the lacI gene of Escherichia coli.
P Quillardet, D Boscus, E Touati, M Hofnung
Index: Mutat. Res. 422(2) , 237-45, (1998)
Full Text: HTML
Abstract
DNA adducts that block replication, induced in vivo by the 5-nitrofuran derivative R7000 (7-methoxy-2-nitronaphtho[2, 1-b]-furan) were mapped, at nucleotide resolution, in a region of the lacI gene of Escherichia coli, using a reiterative primer extension assay [D. Chandrasekhar, B. Van Houten, High resolution mapping of UV-induced photoproducts in the Escherichia coli lacI gene: inefficient repair of the non-transcribed strand correlates with high mutation frequency, J. Mol. Biol., 1994, Vol. 238, pp. 319-332]. It was found that R7000 induced a broad spectrum of low frequency replication blocks rather than particular hot spots in a limited number of particular targets. Most of these replication blocks were observed at G nucleotides, and most of G nucleotides present in the DNA sequence, if not all, constituted a possible target for the chemical attack of the compound. In addition, a large part of replication blocks observed at A, C or T could also reflect a replication block at the 3' or 5' nucleotide flanking a guanosine-DNA adduct. Only a very small number of replication blocks could be observed at A, C or T nucleotides non-adjacent to a G. These results show that, guanosine-DNA adducts are the main DNA lesions that block replication induced by R7000 in E. coli and suggests a strong reactivity of the genotoxic species generated in vivo by R7000 with the G nucleotidic targets. From 26 R7000-induced mutations previously mapped in this region [E. Touati, E. Krin, P. Quillardet, M. Hofnung, 7-methoxy-2-nitronaphto[2,1-b]furan (R7000)-induced mutation spectrum in the lacI gene of Escherichia coli: influence of SOS mutagenesis, Carcinogenesis, 1996, Vol. 17, pp. 2543-2550.], 22 (85%) occurred at GC base pairs at which termination products were observed. The other mutagenic events involved AT base pairs adjacent to a G nucleotide forming a replication block. Thus all mutagenic events occurred at, or adjacent to, a G nucleotide forming a replication block. Although it could not be excluded that some mutagenic events are due to undetected DNA lesions that do not block replication, these results strongly suggest that guanosine-DNA adducts that block DNA replication are responsive for a large part of the mutagenic events generated by R7000. The powerful capacity of R7000 to form adducts at most of the guanosine residues in a DNA sequence may account for at least part of its very potent genotoxic properties.Copyright 1998 Elsevier Science B.V.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
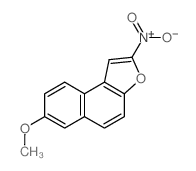 |
7-METHOXY-2-NITRONAPHTHO(2,1-b)FURAN
CAS:75965-74-1 |
C13H9NO4 |
|
Genotoxicity of 2-nitro-7-methoxy-naphtho[2,1-b]furan (R7000...
2002-09-01 [Res. Microbiol. 153(7) , 427-34, (2002)] |
|
First characterization of the mutagenic specificity of 7-met...
1991-05-01 [Mutat. Res. 248(1) , 85-92, (1991)] |
|
Influence of the uvr-dependent nucleotide excision repair on...
1996-10-28 [Mutat. Res. 358(1) , 113-22, (1996)] |
|
Mutagenic properties of a nitrofuran, 7-methoxy-2-nitronapht...
2000-10-31 [Mutat. Res. 470(2) , 177-88, (2000)] |
|
Comparison of the carcinogenic effects of two 2-nitro-naphth...
1987-04-01 [Cancer Lett. 35(1) , 59-64, (1987)] |