Comparison of binding sites in DNA for berenil, netropsin and distamycin. A footprinting study.
J Portugal, M J Waring
Index: Eur. J. Biochem. 167 , 281, (1987)
Full Text: HTML
Abstract
Techniques of DNase I and micrococcal nuclease footprinting have been used to compare the binding sites for berenil, netropsin and distamycin on two different DNA fragments. Each ligand binds to the A + T-rich zones which contain clusters of at least four A.T base pairs. Neither guanosine nor cytidine nucleotides appear to be allowed within the A + T-rich runs which constitute the preferred binding sites, although they are sometimes protected from DNase I cleavage in neighbouring regions. Berenil and netropsin share with distamycin the property of causing enhanced rates of cleavage at certain sequences flanking their binding sites. There are significant differences in the concentrations of each ligand required to produce defined patterns of protection, seemingly dependent upon the nature (and possibly the gross base composition) of the piece of DNA being used in the experiment.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
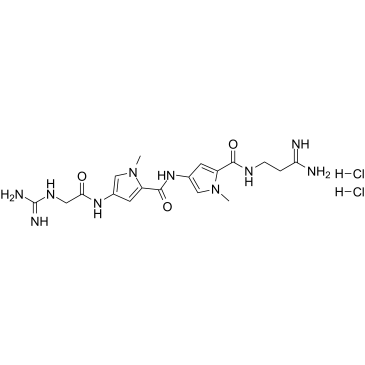 |
Netropsin dihydrochloride
CAS:18133-22-7 |
C18H28Cl2N10O3 |
|
Free energy calculations offer insights into the influence o...
2011-08-01 [J. Comput. Aided Mol. Des. 25(8) , 709-16, (2011)] |
|
Polyamide-scorpion cyclam lexitropsins selectively bind AT-r...
2011-01-01 [PLoS ONE 6(5) , e17446, (2011)] |
|
DNA site-specific N3-adenine methylation targeted to estroge...
2011-09-01 [Bioorg. Med. Chem. 19 , 5093-102, (2011)] |
|
[Estimation of activity of bis-netropsin derivatives based o...
2013-01-01 [Vopr. Virusol. 58(1) , 32-5, (2013)] |
|
A new phenanthroline-oxazine ligand: synthesis, coordination...
2013-03-21 [Chem. Commun. (Camb.) 49(23) , 2341-3, (2013)] |