| Structure | Name/CAS No. | Articles |
|---|---|---|
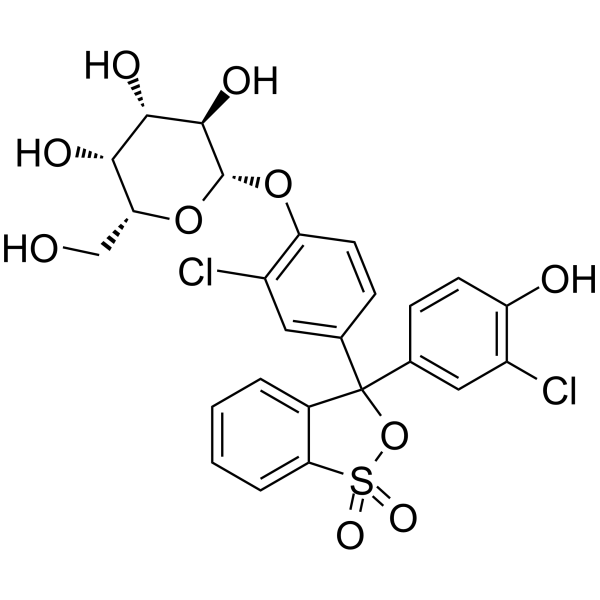 |
chlorophenol red-beta-d-galactopyranosid
CAS:99792-79-7 |
|
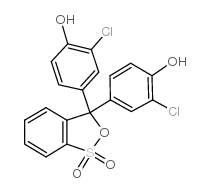 |
Chlorophenol Red
CAS:4430-20-0 |