| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
D-(+)-CELLOTETRAOSE
CAS:38819-01-1 |
|
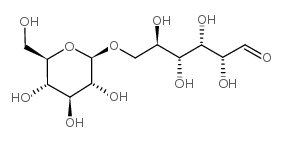 |
β-Gentiobiose
CAS:554-91-6 |
|
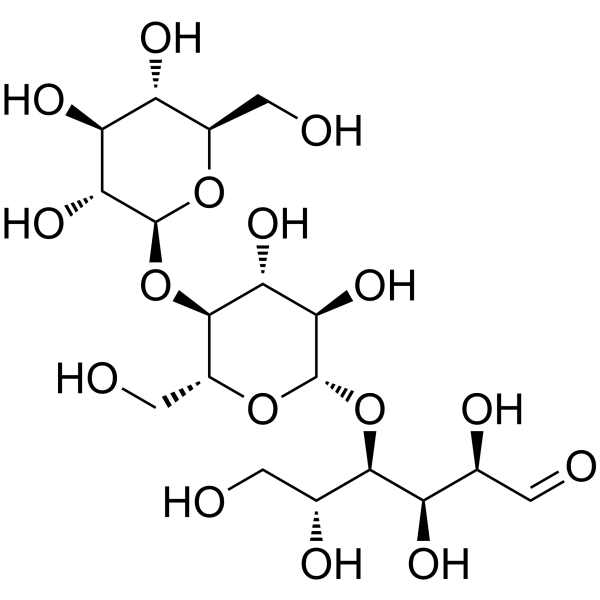 |
D-(+)-CELLOTRIOSE
CAS:33404-34-1 |
|
 |
Maltose
CAS:69-79-4 |