| Structure | Name/CAS No. | Articles |
|---|---|---|
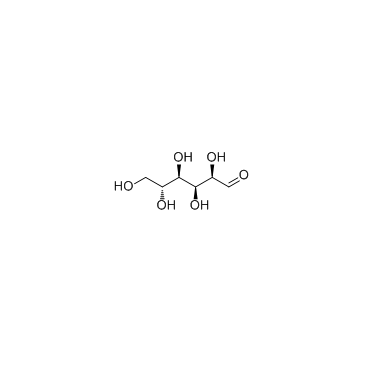 |
D-(+)-Glucose
CAS:50-99-7 |
|
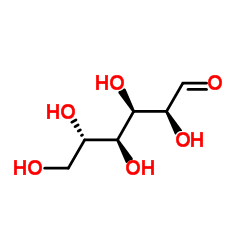 |
L-Glucose
CAS:921-60-8 |
|
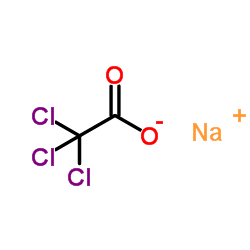 |
Sodium TCA
CAS:650-51-1 |
|
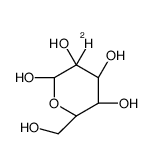 |
(2R,3S,4R,5R)-2-deuterio-2,3,4,5,6-pentahydroxyhexanal
CAS:30737-83-8 |