| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Chloramphenicol
CAS:56-75-7 |
|
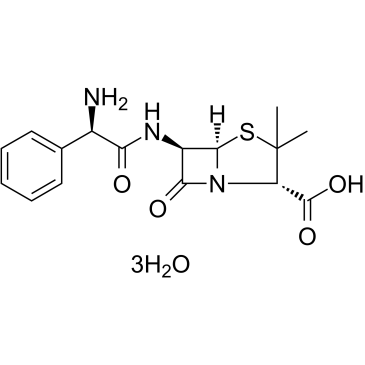 |
Ampicillin Trihydrate
CAS:7177-48-2 |
|
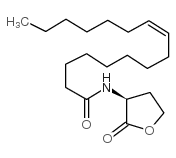 |
2(3H)-Furanone,3-aminodihydro-(8CI,9CI)
CAS:1192-20-7 |
|
 |
Ampicillin
CAS:69-53-4 |