| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
L-(−)-Arabitol
CAS:7643-75-6 |
|
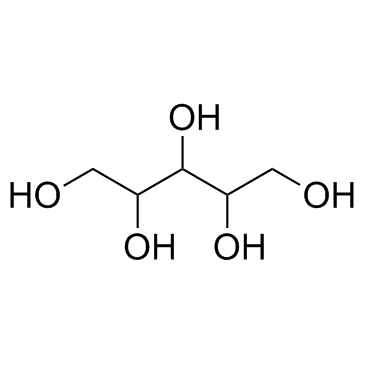 |
D-arabinitol
CAS:488-82-4 |
|
 |
D-Xylulose
CAS:551-84-8 |
|
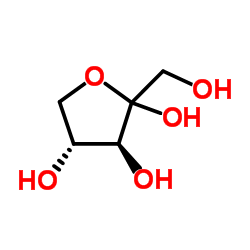 |
L-(+)-Ribulose
CAS:527-50-4 |