| Structure | Name/CAS No. | Articles |
|---|---|---|
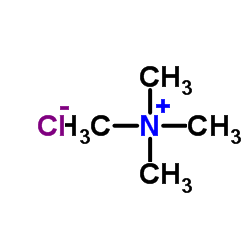 |
Tetramethyl ammonium chloride
CAS:75-57-0 |
|
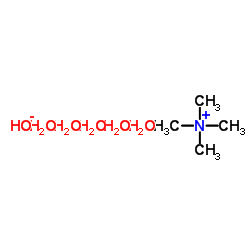 |
Tetramethylammonium Hydroxide Pentahydrate
CAS:10424-65-4 |
|
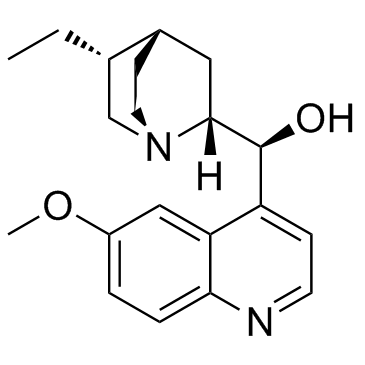 |
Hydroquinidine
CAS:1435-55-8 |
|
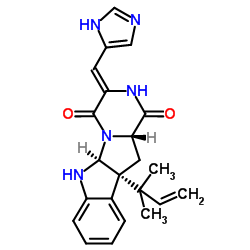 |
isoroquefortine C
CAS:58735-64-1 |
|
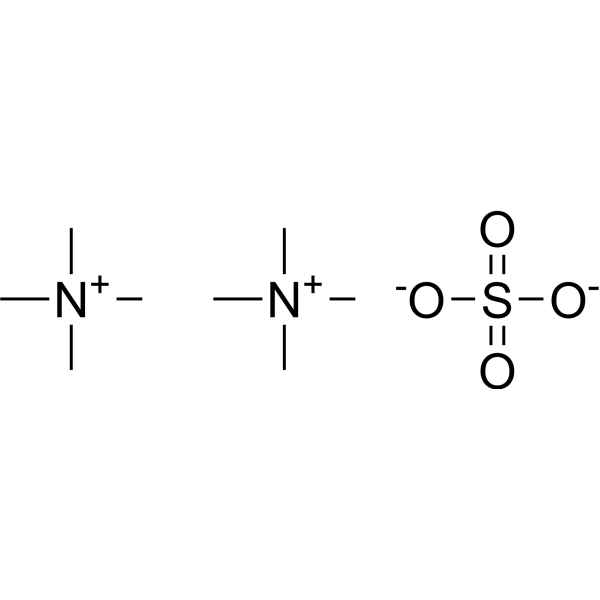 |
Tetramethylammonium Sulfate
CAS:14190-16-0 |
|
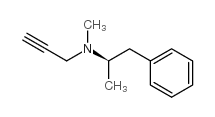 |
selegiline
CAS:14611-51-9 |