| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
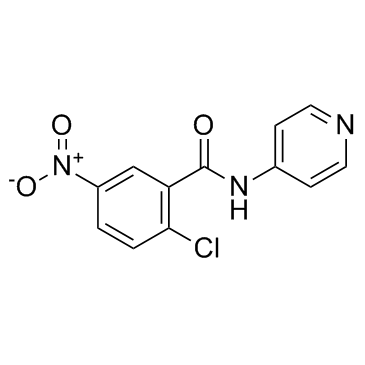 |
T0070907
CAS:313516-66-4 |