| Structure | Name/CAS No. | Articles |
|---|---|---|
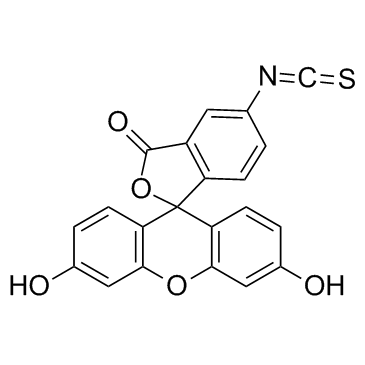 |
Fluorescein isothiocyanate
CAS:3326-32-7 |
|
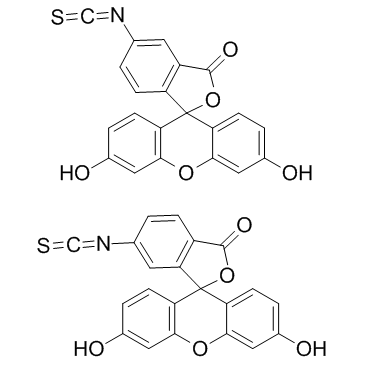 |
fluorescein 5-isothiocyanate
CAS:27072-45-3 |
|
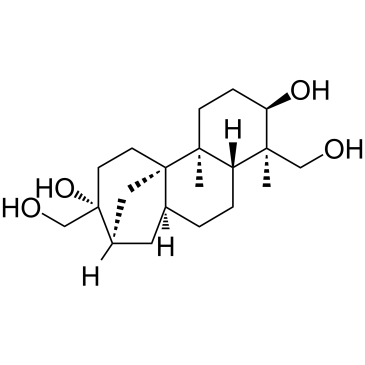 |
(+)-Aphidicolin
CAS:38966-21-1 |
|
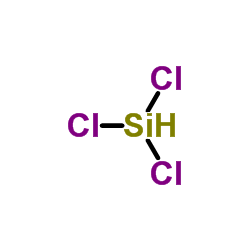 |
Trichlorosilane
CAS:10025-78-2 |