| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
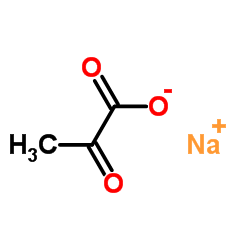 |
Sodium 2-oxopropanoate
CAS:113-24-6 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
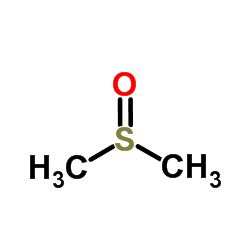 |
Dimethyl sulfoxide
CAS:67-68-5 |
|
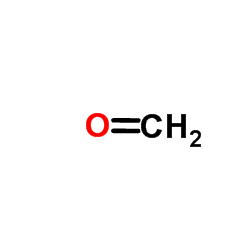 |
Formaldehyde
CAS:50-00-0 |
|
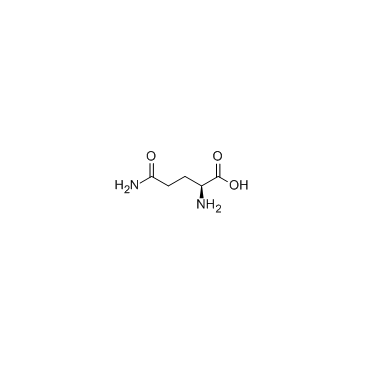 |
L-Glutamine
CAS:56-85-9 |
|
 |
Retinoic acid
CAS:302-79-4 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
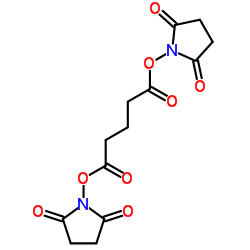 |
DSG Crosslinker
CAS:79642-50-5 |
|
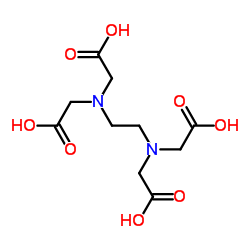 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |