| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sucrose
CAS:57-50-1 |
|
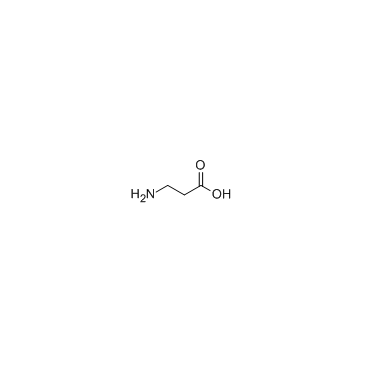 |
beta-Alanine
CAS:107-95-9 |
|
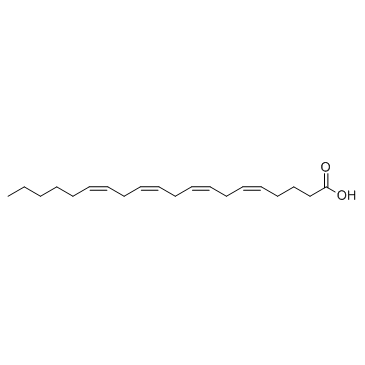 |
Arachidonic acid
CAS:506-32-1 |
|
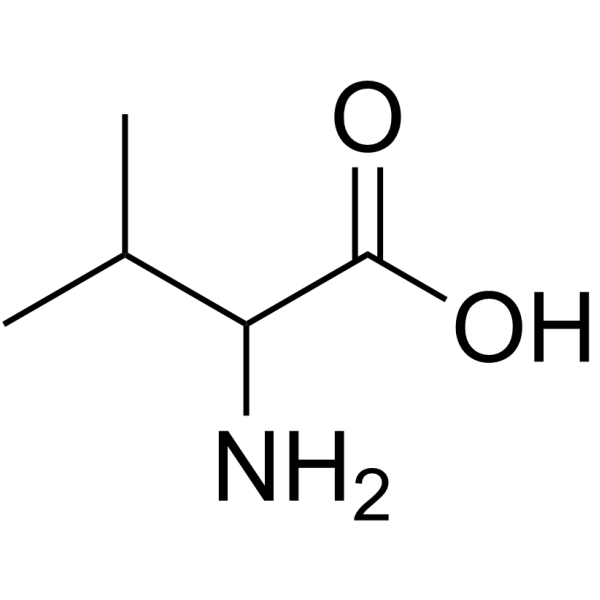 |
DL-Valine
CAS:516-06-3 |
|
 |
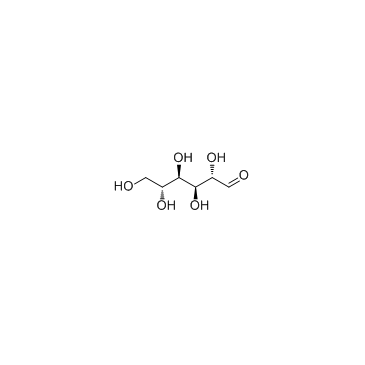
Fructose
CAS:57-48-7 |
|
 |
D-Mannose
CAS:3458-28-4 |
|
 |
L-Isoleucine
CAS:73-32-5 |
|
 |
DL-Serine
CAS:302-84-1 |
|
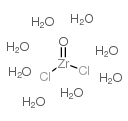 |
Zirconyl chloride octahydrate
CAS:13520-92-8 |
|
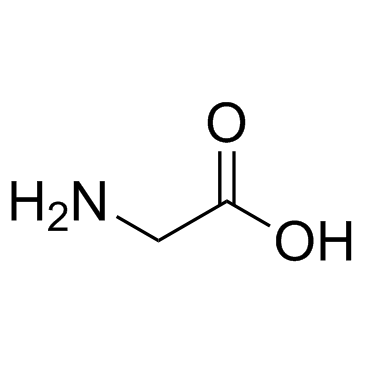 |
Glycine
CAS:56-40-6 |