| Structure | Name/CAS No. | Articles |
|---|---|---|
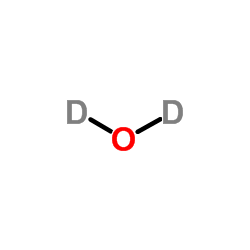 |
Heavy water
CAS:7789-20-0 |
|
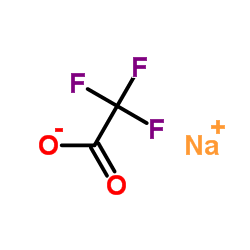 |
Sodium perfluoroacetate
CAS:2923-18-4 |
|
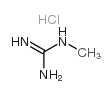 |
1-Methylguanidine hydrochloride
CAS:21770-81-0 |