| Structure | Name/CAS No. | Articles |
|---|---|---|
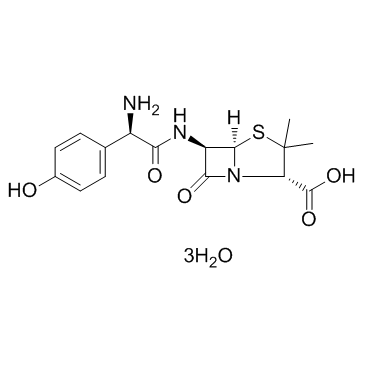 |
Amoxicillin Trihydrate
CAS:61336-70-7 |
|
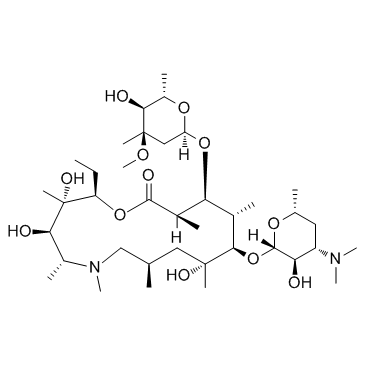 |
Azithromycin
CAS:83905-01-5 |
|
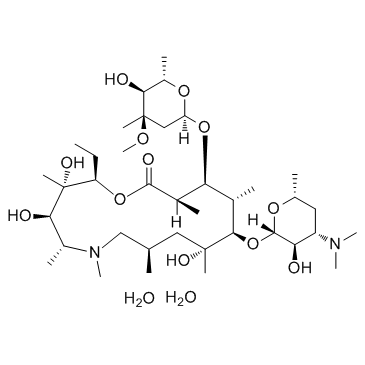 |
Azithromycin Dihydrate
CAS:117772-70-0 |
|
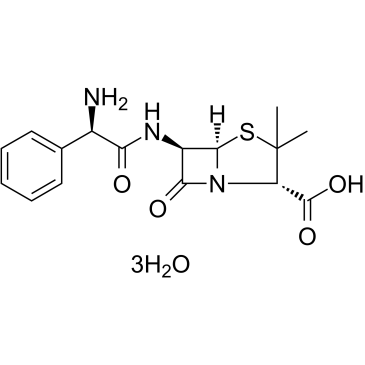 |
Ampicillin Trihydrate
CAS:7177-48-2 |
|
 |
Ampicillin
CAS:69-53-4 |
|
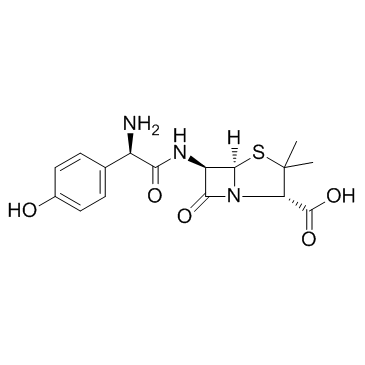 |
Amoxicillin
CAS:26787-78-0 |