| Structure | Name/CAS No. | Articles |
|---|---|---|
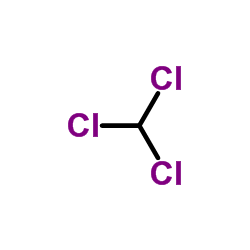 |
Chloroform
CAS:67-66-3 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Ethanol
CAS:64-17-5 |
|
 |
sodium dodecyl sulfate
CAS:151-21-3 |
|
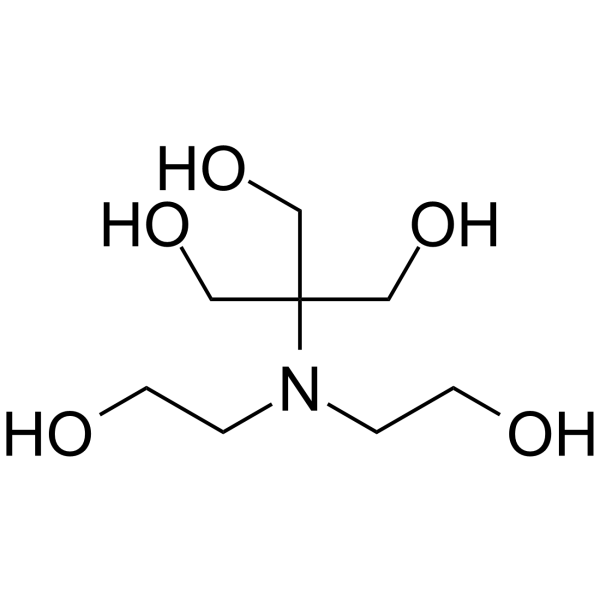 |
Bis-tris methane
CAS:6976-37-0 |
|
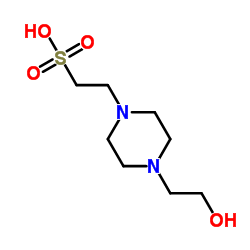 |
HEPES
CAS:7365-45-9 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
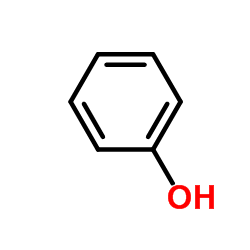 |
Phenol
CAS:108-95-2 |
|
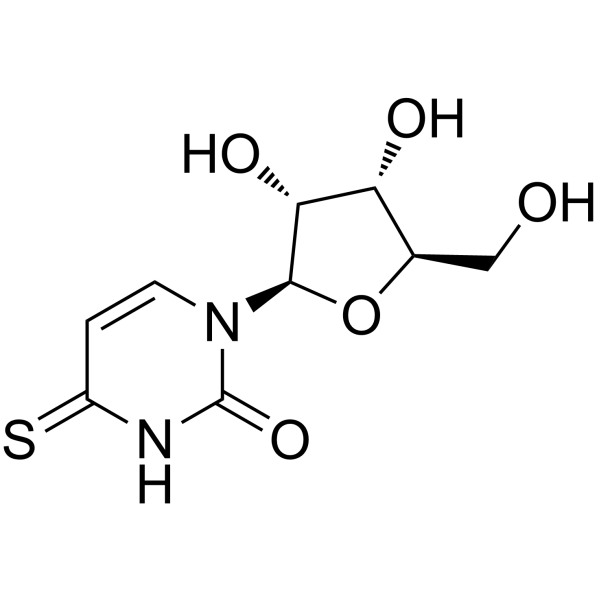 |
4-thiouridine
CAS:13957-31-8 |
|
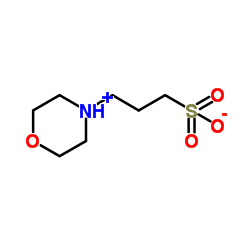 |
MOPS
CAS:1132-61-2 |