Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics.
Annika C Mosier, Nicholas B Justice, Benjamin P Bowen, Richard Baran, Brian C Thomas, Trent R Northen, Jillian F Banfield
文献索引:MBio 4 , e00484-12, (2013)
全文:HTML全文
摘要
Microorganisms grow under a remarkable range of extreme conditions. Environmental transcriptomic and proteomic studies have highlighted metabolic pathways active in extremophilic communities. However, metabolites directly linked to their physiology are less well defined because metabolomics methods lag behind other omics technologies due to a wide range of experimental complexities often associated with the environmental matrix. We identified key metabolites associated with acidophilic and metal-tolerant microorganisms using stable isotope labeling coupled with untargeted, high-resolution mass spectrometry. We observed >3,500 metabolic features in biofilms growing in pH ~0.9 acid mine drainage solutions containing millimolar concentrations of iron, sulfate, zinc, copper, and arsenic. Stable isotope labeling improved chemical formula prediction by >50% for larger metabolites (>250 atomic mass units), many of which were unrepresented in metabolic databases and may represent novel compounds. Taurine and hydroxyectoine were identified and likely provide protection from osmotic stress in the biofilms. Community genomic, transcriptomic, and proteomic data implicate fungi in taurine metabolism. Leptospirillum group II bacteria decrease production of ectoine and hydroxyectoine as biofilms mature, suggesting that biofilm structure provides some resistance to high metal and proton concentrations. The combination of taurine, ectoine, and hydroxyectoine may also constitute a sulfur, nitrogen, and carbon currency in the communities.Microbial communities are central to many critical global processes and yet remain enigmatic largely due to their complex and distributed metabolic interactions. Metabolomics has the possibility of providing mechanistic insights into the function and ecology of microbial communities. However, our limited knowledge of microbial metabolites, the difficulty of identifying metabolites from complex samples, and the inability to link metabolites directly to community members have proven to be major limitations in developing advances in systems interactions. Here, we show that combining stable-isotope-enabled metabolomics with genomics, transcriptomics, and proteomics can illuminate the ecology of microorganisms at the community scale.
相关化合物
| 结构式 | 名称/CAS号 | 分子式 | 全部文献 |
|---|---|---|---|
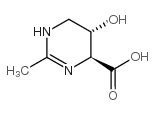 |
羟基四氢嘧啶
CAS:165542-15-4 |
C6H10N2O3 |
|
Synthesis of 5-hydroxyectoine from ectoine: crystal structur...
2010-01-01 [PLoS ONE 5 , e10647, (2010)] |
|
Evaluation of 3-hydroxybutyrate as an enzyme-protective agen...
2016-02-01 [Appl. Microbiol. Biotechnol. 100 , 1365-76, (2016)] |
|
K. Lippert, E.A. Galinski
[Appl. Microbiol. Biotechnol. 37 , 61, (1992)] |
|
T. Sauer et al.
[GIT Labor-Fachzeit. 39 , 892, 894, (1995)] |
|
M.D. Vangala et al.
[Bioforum 23 , 760, (2000)] |