| 结构式 | 名称/CAS号 | 全部文献 |
|---|---|---|
 |
DL-赖氨酸
CAS:70-54-2 |
|
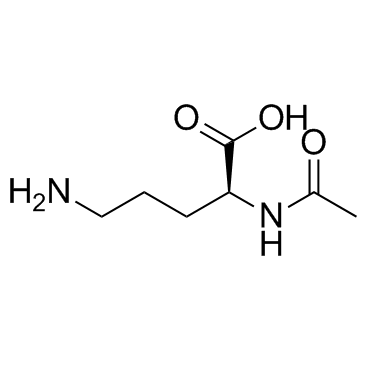 |
N-alpha-乙酰基-L-鸟氨酸
CAS:6205-08-9 |
|
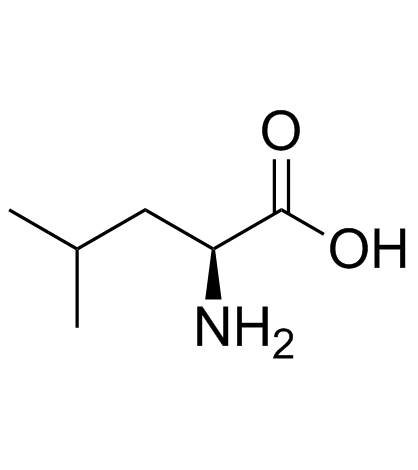 |
L-亮氨酸
CAS:61-90-5 |
|
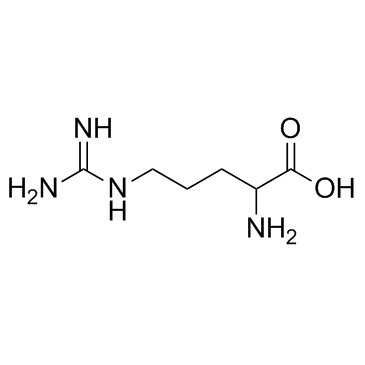 |
DL-精氨酸
CAS:7200-25-1 |
|
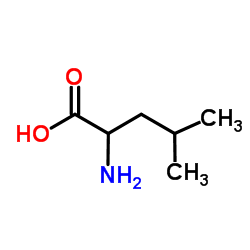 |
(±)-氨基-4-甲基戊酸
CAS:328-39-2 |