| 结构式 | 名称/CAS号 | 全部文献 |
|---|---|---|
 |
神经氨酸酶 来源于产气荚膜梭菌
CAS:9001-67-6 |
|
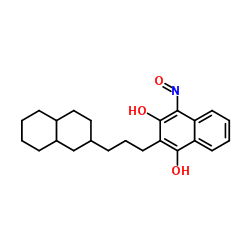 |
酯酶 来源于猪肝脏
CAS:9016-18-6 |