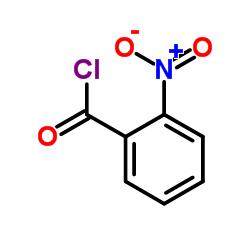
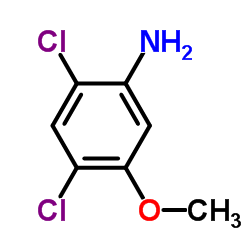
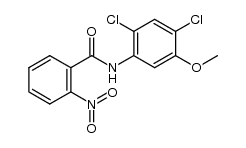
338967-87-6
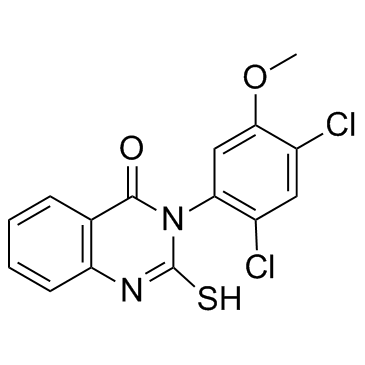
| Name | 3-(2,4-dichloro-5-methoxyphenyl)-2-sulfanylidene-1H-quinazolin-4-one |
|---|---|
| Synonyms |
4(1H)-Quinazolinone, 3-(2,4-dichloro-5-methoxyphenyl)-2,3-dihydro-2-thioxo-
HMS575G09 3-(2,4-Dichloro-5-methoxyphenyl)-2-thioxo-2,3-dihydro-4(1H)-quinazolinone Mdivi-1 3-(2,4-Dichloro-5-methoxyphenyl)-2-sulfanylquinazolin-4(3H)-one 3-(2,4-Dichloro-5-methoxyphenyl)-2,3-dihydro-2-thioxo-4(1H)-quinazolinone Mitochondrial division inhibitor 1 |
| Description | Mdivi-1 is a selective dynamin-related protein 1 (Drp1) inhibitor. |
|---|---|
| Related Catalog | |
| In Vitro | Mdivi-1 inhibits Dnm1 GTPase activity in a dose-dependent manner, with an estimated EC50 of 1-10 μM. Mdivi-1 increases the apparent K0.5 for GTP, lowers the apparent Vmax for GTP hydrolysis, and causes an increase in the Hill coefficient observed for GTP in the Dnm1 GTP hydrolysis reaction[1]. Cells treated with mdivi-1 display decreased cytochrome c release and a reduced rate of phosphatidylserine exposure on their surface following apoptosis induction, consistent with an inhibition of apoptosis and with previous studies using other strategies to compromise DRP1 activity[2]. Mdivi-1 results in apoptotic cell death in ischemic retina[3]. |
| In Vivo | The mitochondrial division DRP, Dnm1, is the target of mdivi-1 in vivo. Mdivi-1 quantitatively blocks GMPPCP- dependent Dnm1 self-assembly in a concentration range similar to its effects on mitochondrial division in vivo[1]. Mdivi-1 (50 mg/kg, i.p.) significantly decreases GFAP protein expression in the normal mouse retina[3]. |
| Kinase Assay | All GTPase assay reactions are started in a 200 μL volume, of which 150 μL is placed into the well of a 96-well plate. Depletion of NADH, as monitored by reading the A340 of the reaction, is measured every 20 s for a total of 40 min using a SpectraMAX 250 96-well plate reader. Spectrophotometric data are transferred to Excel and the measured steady state depletion of NADH over time is converted to protein activity. |
| Cell Assay | YPGlycerol plates are topped with 10 mL YPGlycerol containing 1% low melt agar and 75 μM mdivi-1, and cells are spotted 12 hours later using a 48 well pinning device. After pinning cells, plates are incubated at 24°C or 37°C and imaged using an Eagle Eye II imaging system. |
| References |
| Density | 1.6±0.1 g/cm3 |
|---|---|
| Boiling Point | 522.5±60.0 °C at 760 mmHg |
| Melting Point | 289 °C |
| Molecular Formula | C15H10Cl2N2O2S |
| Molecular Weight | 353.223 |
| Flash Point | 269.8±32.9 °C |
| Exact Mass | 351.984009 |
| PSA | 82.92000 |
| LogP | 3.85 |
| Vapour Pressure | 0.0±1.4 mmHg at 25°C |
| Index of Refraction | 1.729 |
| Storage condition | Store at -20°C |
| Water Solubility | DMSO: >20mg/mL |
| Hazard Codes | Xi |
|---|---|
| RIDADR | NONH for all modes of transport |
|
~% 
338967-87-6 |
| Literature: Matsumoto, Yotaro; Noguchi-Yachide, Tomomi; Nakamura, Masaharu; Mita, Yusuke; Numadate, Akiyoshi; Hashimoto, Yuichi Heterocycles, 2012 , vol. 86, # 2 p. 1449 - 1463 |
|
~% 
338967-87-6 |
| Literature: Matsumoto, Yotaro; Noguchi-Yachide, Tomomi; Nakamura, Masaharu; Mita, Yusuke; Numadate, Akiyoshi; Hashimoto, Yuichi Heterocycles, 2012 , vol. 86, # 2 p. 1449 - 1463 |
|
~% 
338967-87-6 |
| Literature: Matsumoto, Yotaro; Noguchi-Yachide, Tomomi; Nakamura, Masaharu; Mita, Yusuke; Numadate, Akiyoshi; Hashimoto, Yuichi Heterocycles, 2012 , vol. 86, # 2 p. 1449 - 1463 |
| Precursor 3 | |
|---|---|
| DownStream 0 | |