Structure and cooperativity of a T-state mutant of histidine decarboxylase from Lactobacillus 30a.
Scott Worley, Elisabeth Schelp, Arthur F Monzingo, Stephen Ernst, Jon D Robertus
Index: Proteins 46(3) , 321-9, (2002)
Full Text: HTML
Abstract
Histidine decarboxylase (HDC) from Lactobacillus 30a converts histidine to histamine, a process that enables the bacteria to maintain the optimum pH range for cell growth. HDC is regulated by pH; it is active at low pH and inactive at neutral to alkaline pH. The X-ray structure of HDC at pH 8 revealed that a helix was disordered, resulting in the disruption of the substrate-binding site. The HDC trimer has also been shown to exhibit cooperative kinetics at neutral pH, that is, histidine can trigger a T-state to R-state transition. The D53,54N mutant of HDC has an elevated Km, even at low pH, indicating that the enzyme assumes the low activity T-state. We have solved the structures of the D53,54N mutant at low pH, with and without the substrate analog histidine methyl ester (HME) bound. Structural analysis shows that the apo-D53,54N mutant is in the inactive or T-state and that binding of the substrate analog induces the enzyme to adopt the active or R-state. A mechanism for the cooperative transition is proposed.Copyright 2002 Wiley-Liss, Inc.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
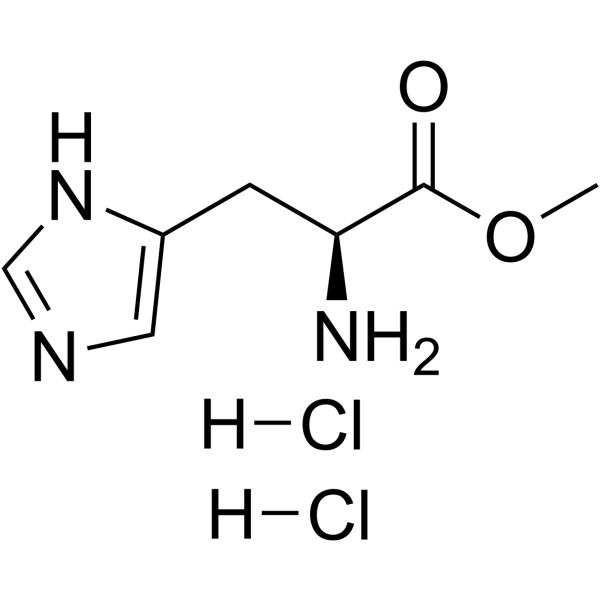 |
H-His-OMe.2HCl
CAS:7389-87-9 |
C7H13Cl2N3O2 |
|
Preparation and use of poly(hydroxyethyl methacrylate) cryog...
2013-02-01 [J. Food Sci. 78(2) , E238-43, (2013)] |
|
A proximal pair of positive charges provides the dominant li...
2012-04-01 [Biochem. J. 443(1) , 65-73, (2012)] |
|
The biological activity of hydrogen peroxide. IV. Enhancemen...
1988-03-01 [Mutat. Res. 198(1) , 233-40, (1988)] |
|
Carbonic anhydrase activators: gold nanoparticles coated wit...
2011-03-10 [J. Med. Chem. 54(5) , 1170-7, (2011)] |
|
Gas-phase structure of protonated histidine and histidine me...
2005-09-22 [J. Phys. Chem. A 109(37) , 8329-35, (2005)] |