Identification of A-minor tertiary interactions within a bacterial group I intron active site by 3-deazaadenosine interference mapping.
Juliane K Soukup, Noriaki Minakawa, Akira Matsuda, Scott A Strobel
Index: Biochemistry 41(33) , 10426-38, (2002)
Full Text: HTML
Abstract
The A-minor motifs appear to be the most ubiquitous helix packing elements within RNA tertiary structures. These motifs have been identified throughout the ribosome and almost every other tertiary-folded RNA for which structural information is available. These motifs utilize the packing of the donor adenosine's N1, N3, and/or 2'-OH against the 2'-OHs and minor groove edge of the acceptor base pair. The ability to identify biochemically which adenosines form A-minor motifs and which base pairs they contact is an important experimental objective. Toward this goal, we report the synthesis and transcriptional incorporation of 5'-O-(1-thio)-3-deazaadenosine triphosphate and its use in Nucleotide Analogue Interference Mapping (NAIM) and Nucleotide Analogue Interference Suppression (NAIS). This analogue makes it possible for the first time to explore the functional importance of the N3 imino group of adenosine in RNA polymers. Interference analysis of the group I self-splicing introns from Tetrahymena and Azoarcus indicates that A-minor motifs are integral to the helix packing interactions that define the 5'-splice site of the intron. Specifically, Azoarcus A58 in the J4/5 region contacts the G.U wobble pair at the cleavage site in the P1 helix, and Azoarcus A167 in the J8/7 region contacts the C13-G37 base pair in the P2 helix. Both of these structural features are conserved between the eukaryotic and bacterial introns. These results suggest that nucleotide analogue interference patterns can identify and distinguish A-minor interactions in RNA tertiary structure, particularly the most prevalent type I and type II varieties. Furthermore, clustering of 3-deazaadenosine interferences is suggestive of A patches, in which a series of consecutive A-minor motifs mediate helix packing. Biochemical identification of these interactions may provide valuable constraints for RNA structure prediction.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
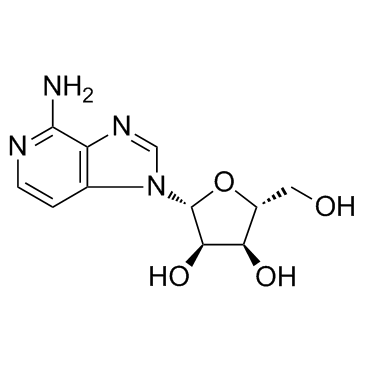 |
3-Deazaadenosine
CAS:6736-58-9 |
C11H14N4O4 |
|
Disruption of choline methyl group donation for phosphatidyl...
2002-05-10 [J. Biol. Chem. 277(19) , 17217-25, (2002)] |
|
Activation of caspase-8 in 3-deazaadenosine-induced apoptosi...
2001-12-31 [Exp. Mol. Med. 33(4) , 284-92, (2001)] |
|
Dietary supplementation with 3-deaza adenosine, N-acetyl cys...
2004-01-01 [Neuromolecular Med. 6(2-3) , 93-103, (2004)] |
|
3-Deazaadenosine inhibits vasa vasorum neovascularization in...
2009-01-01 [Atherosclerosis 202(1) , 103-10, (2009)] |
|
Protein methylation activates reconstituted ryanodine recept...
2004-01-01 [J. Vasc. Res. 41(3) , 229-40, (2004)] |