| Structure | Name/CAS No. | Articles |
|---|---|---|
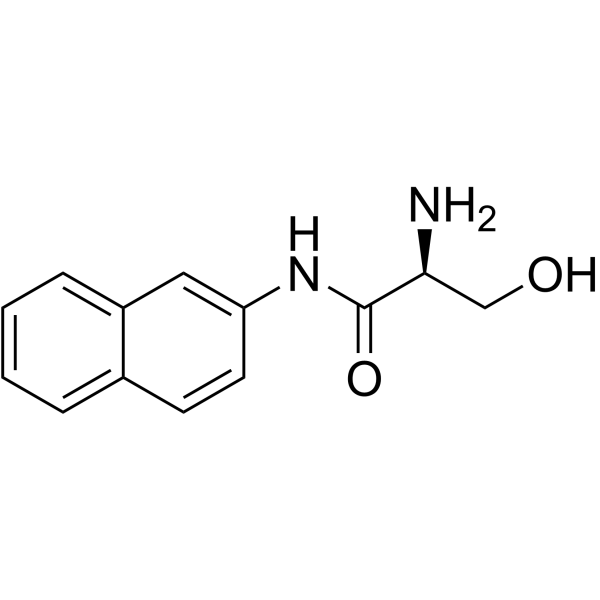 |
L-SERINE BETA-NAPHTHYLAMIDE
CAS:888-74-4 |
| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
L-SERINE BETA-NAPHTHYLAMIDE
CAS:888-74-4 |