| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
Trimethoprim
CAS:738-70-5 |
|
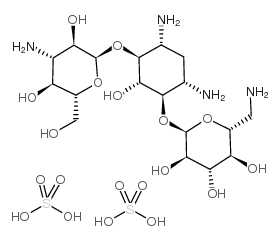 |
kanamycin acid sulfate
CAS:64013-70-3 |
|
 |
Chloramphenicol
CAS:56-75-7 |