[Oligoglycine surface structures: molecular dynamics simulation].
O A Gus'kova, P G Khalatur, A R Khokhlov, A A Chinarev, S V Tsygankova, N V Bovin
Index: Bioorg. Khim. 36(5) , 622-9, (2010)
Full Text: HTML
Abstract
The full-atomic molecular dynamics (MD) simulation of adsorption mode for diantennary oligoglycines [H-Gly4-NH(CH2)5]2 onto graphite and mica surface is described. The resulting structure of adsorption layers is analyzed. The peptide second structure motives have been studied by both STRIDE (structural identification) and DSSP (dictionary of secondary structure of proteins) methods. The obtained results confirm the possibility of polyglycine II (PGII) structure formation in diantennary oligoglycine (DAOG) monolayers deposited onto graphite surface, which was earlier estimated based on atomic-force microscopy measurements.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
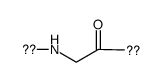 |
Polyglycine
CAS:25718-94-9 |
C2H3NO |
|
Aggregation of capped hexaglycine strands into hydrogen-bond...
2011-02-17 [J. Phys. Chem. B 115 , 1562-1570, (2011)] |
|
Catalytic oxidation of uric acid at the polyglycine chemical...
1997-08-01 [Analyst 122 , 839-841, (1997)] |
|
One-pot electrosynthesis of polyglycine-like thin film on pl...
2010-06-30 [Talanta 82 , 417-421, (2010)] |
|
Communications: Is quantum chemical treatment of biopolymers...
2010-06-21 [J. Chem. Phys. 132(23) , 231101, (2010)] |
|
Structure of sodiated octa-glycine: IRMPD spectroscopy and m...
2010-05-01 [J. Am. Soc. Mass Spectrom. 21(5) , 728-38, (2010)] |