| Structure | Name/CAS No. | Articles |
|---|---|---|
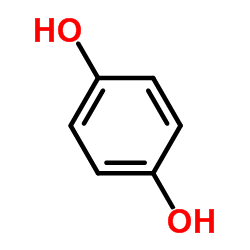 |
Hydroquinone
CAS:123-31-9 |
|
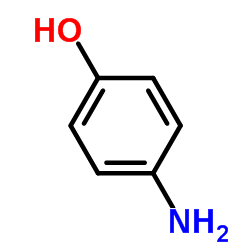 |
4-Aminophenol
CAS:123-30-8 |
|
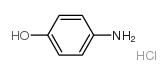 |
|p|-Aminophenol hydrochloride
CAS:51-78-5 |