Passenger deletions generate therapeutic vulnerabilities in cancer.
Florian L Muller, Simona Colla, Elisa Aquilanti, Veronica E Manzo, Giannicola Genovese, Jaclyn Lee, Daniel Eisenson, Rujuta Narurkar, Pingna Deng, Luigi Nezi, Michelle A Lee, Baoli Hu, Jian Hu, Ergun Sahin, Derrick Ong, Eliot Fletcher-Sananikone, Dennis Ho, Lawrence Kwong, Cameron Brennan, Y Alan Wang, Lynda Chin, Ronald A DePinho
Index: Nature 488(7411) , 337-42, (2012)
Full Text: HTML
Abstract
Inactivation of tumour-suppressor genes by homozygous deletion is a prototypic event in the cancer genome, yet such deletions often encompass neighbouring genes. We propose that homozygous deletions in such passenger genes can expose cancer-specific therapeutic vulnerabilities when the collaterally deleted gene is a member of a functionally redundant family of genes carrying out an essential function. The glycolytic gene enolase 1 (ENO1) in the 1p36 locus is deleted in glioblastoma (GBM), which is tolerated by the expression of ENO2. Here we show that short-hairpin-RNA-mediated silencing of ENO2 selectively inhibits growth, survival and the tumorigenic potential of ENO1-deleted GBM cells, and that the enolase inhibitor phosphonoacetohydroxamate is selectively toxic to ENO1-deleted GBM cells relative to ENO1-intact GBM cells or normal astrocytes. The principle of collateral vulnerability should be applicable to other passenger-deleted genes encoding functionally redundant essential activities and provide an effective treatment strategy for cancers containing such genomic events.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
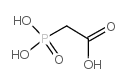 |
Acetic acid,2-phosphono-
CAS:4408-78-0 |
C2H5O5P |
|
Equine Herpesvirus 1 Multiply Inserted Transmembrane Protein...
2015-06-15 [J. Virol. 89 , 6251-63, (2015)] |
|
Epstein-barr virus blocks the autophagic flux and appropriat...
2014-11-01 [J. Virol. 88(21) , 12715-26, (2014)] |
|
Viral semaphorin inhibits dendritic cell phagocytosis and mi...
2015-04-01 [J. Virol. 89(7) , 3630-47, (2015)] |
|
Interaction between the biotin carboxyl carrier domain and t...
2011-11-15 [Biochemistry 50(45) , 9708-23, (2011)] |
|
Structural insight into inositol pyrophosphate turnover.
2013-01-01 [Adv. Biol. Regul. 53(1) , 19-27, (2013)] |