| Structure | Name/CAS No. | Articles |
|---|---|---|
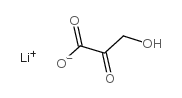 |
BETA-HYDROXYPYRUVIC ACID LITHIUM SALT HYDRATE
CAS:3369-79-7 |
|
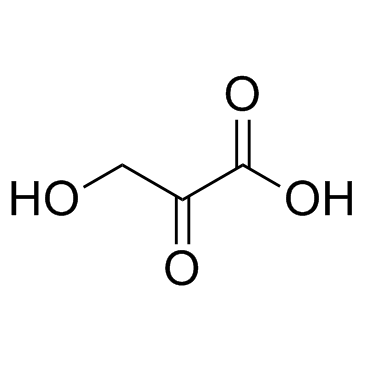 |
Hydroxypyruvic acid
CAS:1113-60-6 |
|
 |
Transketolase from E. coli
CAS:9014-48-6 |