| Structure | Name/CAS No. | Articles |
|---|---|---|
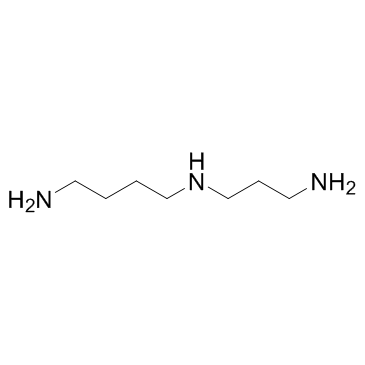 |
spermidine
CAS:124-20-9 |
|
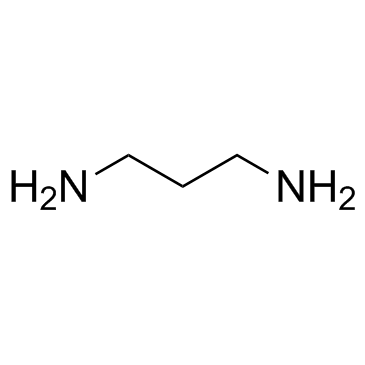 |
1,3-Diaminopropane
CAS:109-76-2 |
|
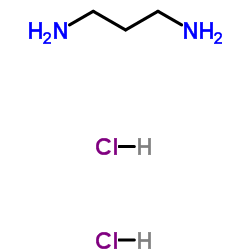 |
1,3-Propanediamine dihydrochloride
CAS:10517-44-9 |