| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
Potassium
CAS:7440-09-7 |
|
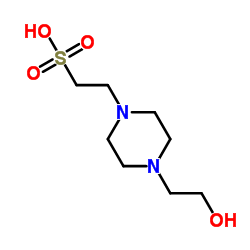 |
HEPES
CAS:7365-45-9 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
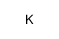 |
potassium hydride
CAS:7693-26-7 |
|
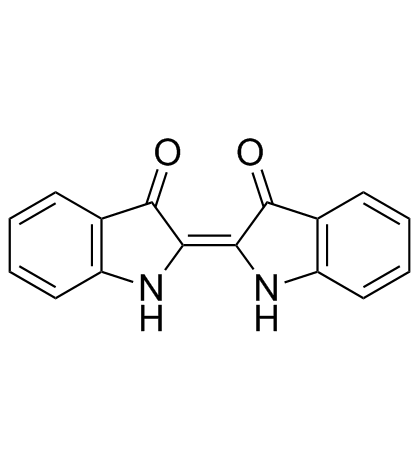 |
Indigo
CAS:482-89-3 |
|
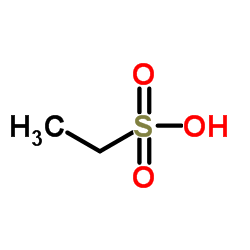 |
Ethanesulfonic acid
CAS:594-45-6 |
|
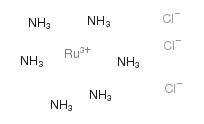 |
Ruthenium(3+) chloride ammoniate (1:3:6)
CAS:14282-91-8 |
|
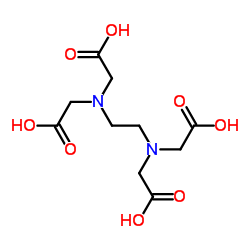 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
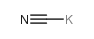 |
POTASSIUM CYANIDE
CAS:151-50-8 |