| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
sodium chloride
CAS:7647-14-5 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
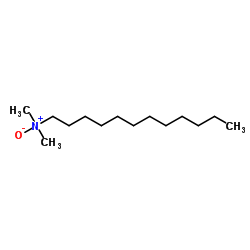 |
N-Dodecyl-N,N-dimethylamine oxide
CAS:1643-20-5 |