| Structure | Name/CAS No. | Articles |
|---|---|---|
 |
L(-)-Tryptophan
CAS:73-22-3 |
|
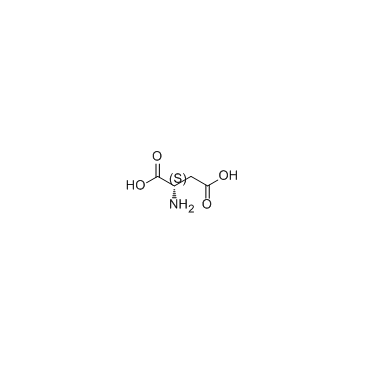 |
L-Aspartic acid
CAS:56-84-8 |
|
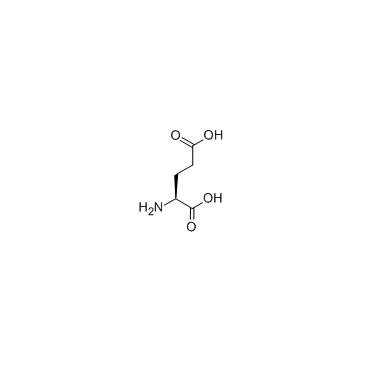 |
L-glutamic acid
CAS:56-86-0 |
|
 |
Phosphorylase b
CAS:9012-69-5 |
|
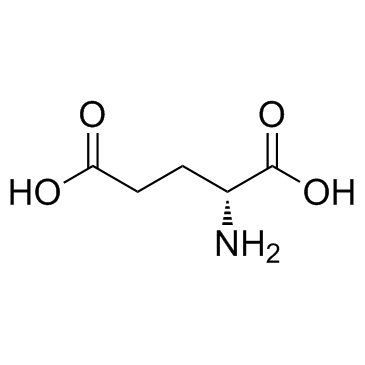 |
D(-)-Glutamic acid
CAS:6893-26-1 |
|
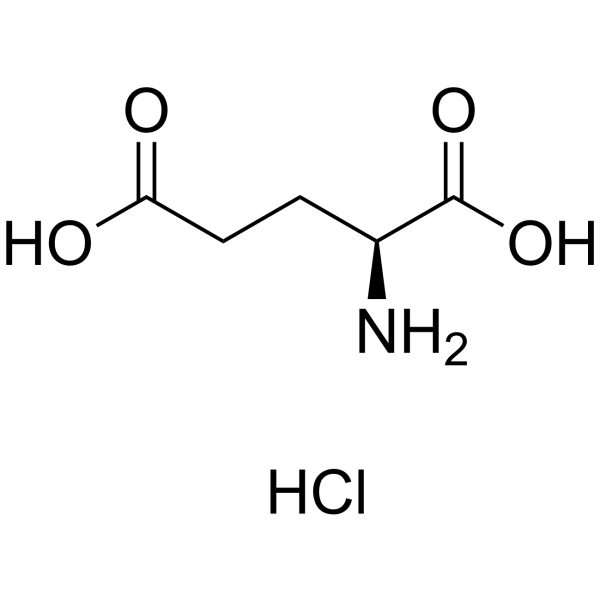 |
L-Glutamic acid:Hcl (17O4)
CAS:138-15-8 |
|
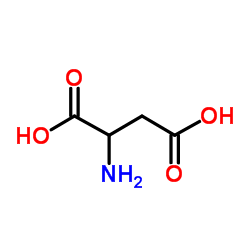 |
DL-Aspartic Acid
CAS:617-45-8 |
|
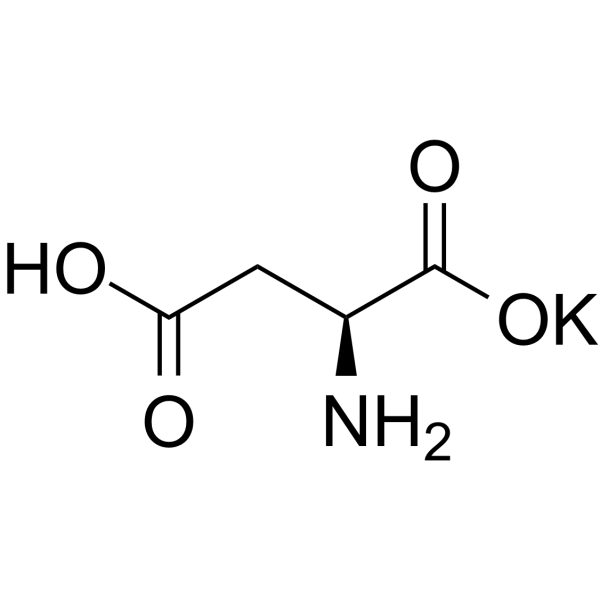 |
Butanedioate, 2-amino-, potassium salt (1:1)
CAS:1115-63-5 |
|
 |
Enterobactin from Escherichia coli
CAS:28384-96-5 |