| Structure | Name/CAS No. | Articles |
|---|---|---|
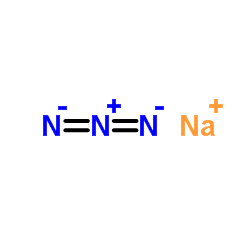 |
Sodium azide
CAS:26628-22-8 |
|
 |
sodium chloride
CAS:7647-14-5 |
|
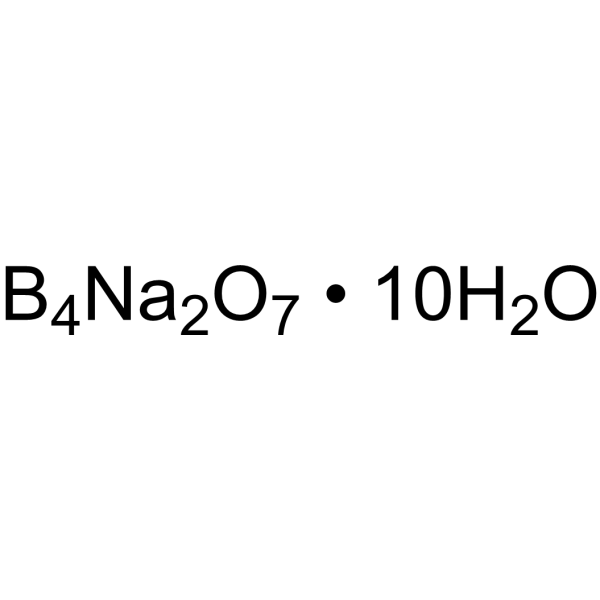 |
Borax
CAS:1303-96-4 |
|
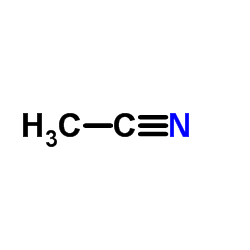 |
Acetonitrile
CAS:75-05-8 |
|
 |
Methanol
CAS:67-56-1 |
|
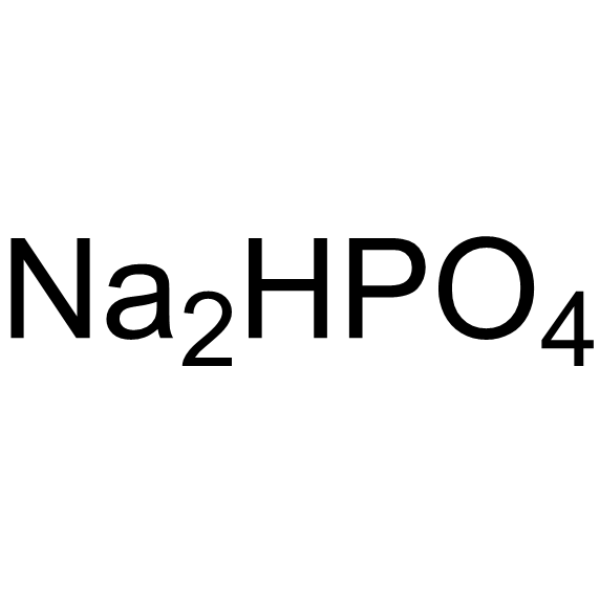 |
Disodium hydrogenorthophosphate
CAS:7558-79-4 |
|
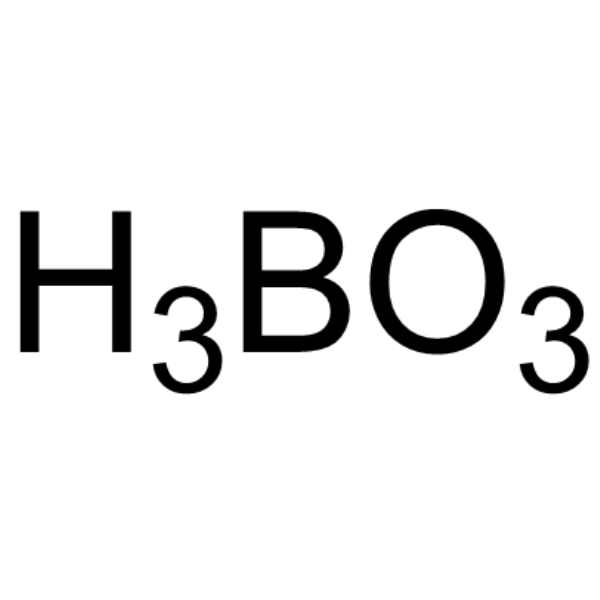 |
Orthoboric acid
CAS:10043-35-3 |
|
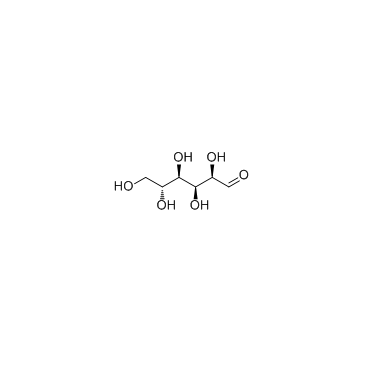 |
D-(+)-Glucose
CAS:50-99-7 |
|
 |
SODIUM CHLORIDE-35 CL
CAS:20510-55-8 |
|
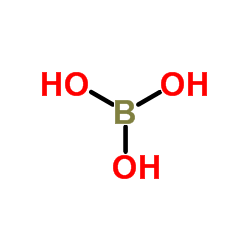 |
Boric acid-11B
CAS:13813-78-0 |