| Structure | Name/CAS No. | Articles |
|---|---|---|
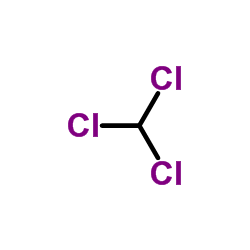 |
Chloroform
CAS:67-66-3 |
|
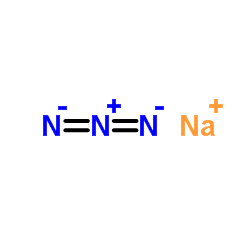 |
Sodium azide
CAS:26628-22-8 |
|
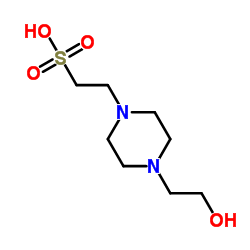 |
HEPES
CAS:7365-45-9 |
|
 |
Carbon
CAS:7440-44-0 |
|
 |
carbon black
CAS:1333-86-4 |
|
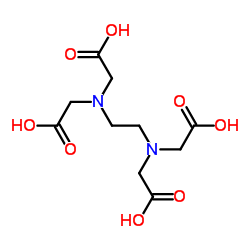 |
Ethylenediaminetetraacetic acid
CAS:60-00-4 |
|
 |
METHANE
CAS:74-82-8 |
|
 |
Succinic anhydride
CAS:108-30-5 |
|
 |
Velcorin
CAS:4525-33-1 |