Altering residues N125 and D149 impacts sugar effector binding and allosteric parameters in Escherichia coli lactose repressor.
Jia Xu, Shirley Liu, Mingzhi Chen, Jianpeng Ma, Kathleen S Matthews
Index: Biochemistry 50(42) , 9002-13, (2011)
Full Text: HTML
Abstract
Lactose repressor protein (LacI), a negative transcriptional regulator in Escherichia coli, relies on an allosteric conformational change for its function. The LacI effector isopropyl-β,D-thiogalactoside (IPTG) promotes this allosteric response and engages the side chains of residues N125 and D149 based on the crystallographic structure of LacI·IPTG. Targeted molecular dynamics (TMD) simulations have indicated involvement of these side chains during the protein structural changes in response to inducer binding. To examine this region further, we applied stochastic boundary molecular dynamics (SBMD) simulation and identified a transient interaction between residues N125 and D149. On the basis of these data, we introduced substitutions for either/both residues and analyzed their impact on protein function. The substitutions utilized were alanine to preclude hydrogen bonding or cysteine to allow disulfide bond formation, which was not observed for N125C/D149C. Minimal impacts were observed on operator affinity for all substitutions, but D149C, N125A/D149A, and N125C/D149C bound to IPTG with 5-8-fold lower affinity than wild-type LacI, and exhibited decreased allosteric amplitude (K(RI/O)/K(R/O)). Of interest, the double mutants did not exhibit an allosteric response to an alternate inducer, 2-phenylethyl-β,D-galactoside (PhEG), despite demonstration of PhEG binding. Further, the presence of the anti-inducer, o-nitrophenyl-β,D-fucoside (ONPF), enhanced operator affinity for wild-type LacI and all other mutant proteins examined, but behaved as an inducer for N125A/D149A, decreasing operator binding affinity. These results confirm the role of residues 125 and 149 in ligand binding and allosteric response and illustrate how readily the function of a regulatory protein can be altered.
Related Compounds
| Structure | Name/CAS No. | Molecular Formula | Articles |
|---|---|---|---|
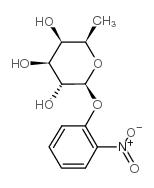 |
2-nitrophenyl β-D-fucoside
CAS:1154-94-5 |
C12H15NO7 |
|
A closer view of the conformation of the Lac repressor bound...
2000-03-01 [Nat. Struct. Biol. 7(3) , 209-14, (2000)] |
|
Thermal denaturation of the core protein of lac repressor.
1985-07-16 [Biochemistry 24(15) , 3842-6, (1985)] |
|
Escherichia coli lac repressor-lac operator interaction and ...
1997-01-10 [J. Mol. Biol. 265(1) , 1-7, (1997)] |
|
Using networks to identify fine structural differences betwe...
2004-08-31 [Biochemistry 43(34) , 10886-95, (2004)] |
|
Ligand interactions with lactose repressor protein and the r...
2007-03-01 [Biophys. Chem. 126(1-3) , 94-105, (2007)] |