| Structure | Name/CAS No. | Articles |
|---|---|---|
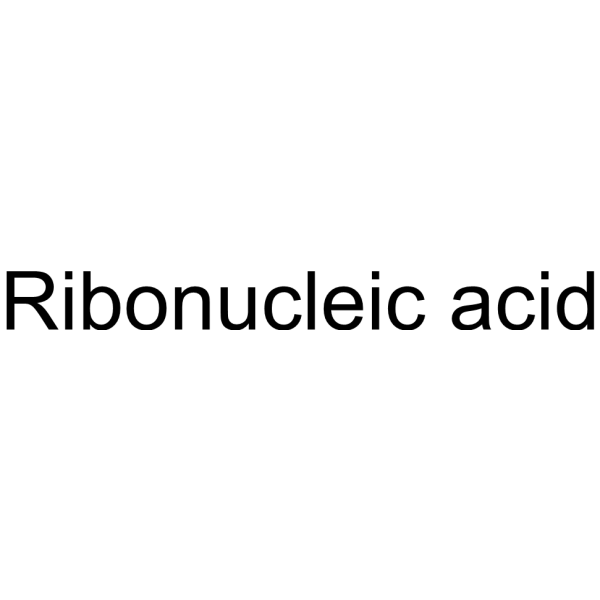 |
Ribonucleic acid
CAS:63231-63-0 |
|
 |
Transfer ribonucleic acids
CAS:9014-25-9 |