| Structure | Name/CAS No. | Articles |
|---|---|---|
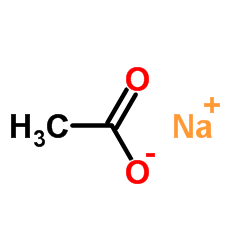 |
Sodium acetate
CAS:127-09-3 |
|
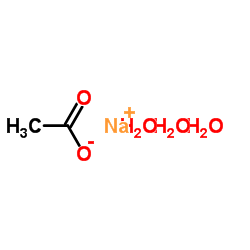 |
sodium acetate trihydrate
CAS:6131-90-4 |