| Structure | Name/CAS No. | Articles |
|---|---|---|
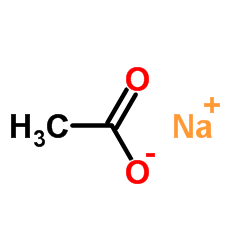 |
Sodium acetate
CAS:127-09-3 |
|
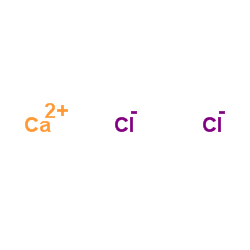 |
Calcium chloride
CAS:10043-52-4 |
|
 |
Erythromycin
CAS:114-07-8 |
|
 |
Potassium (1R,2S,3R,4R,5S,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl phosphate
CAS:129832-03-7 |
|
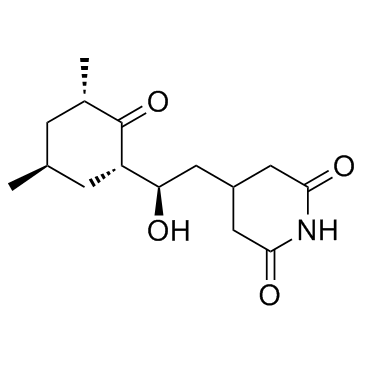 |
Cycloheximide
CAS:66-81-9 |
|
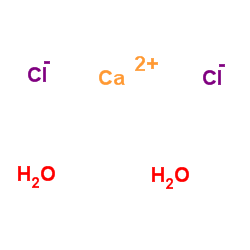 |
calcium chloride dihydrate
CAS:10035-04-8 |
|
 |
Chloramphenicol
CAS:56-75-7 |
|
 |
Inositol
CAS:87-89-8 |